The benefits of this technology are not limited to environmental impact. Through elicitation of red rice meristematic cell cultures, we were able to produce appreciable quantities of secondary metabolites in the culture. When the culture was tested on fibroblasts in vitro, we show a reverse in the age related increase of methylation in the promoter region of genes. This rejuvenation at the epigenetic level may have multiple benefits to the cells and to the skin. Some benefits include anti-aging and the renewal of gene and protein expression levels to that found in younger cells. In essence, the skin, through an application of ReGeniStemTM Red Rice, is pushed to a younger, healthier state. An increased expression of dermatopontin in keratinocytes, besides being a novel finding, implies that ReGeniStemTM Red Rice may be able to firm the skin and improve barrier function.
Philip Ludwig, Senior Research Chemist; Suellen Bennett, Global Manager, Technical Marketing; James V. Gruber, Ph.D., Director of Research & Market Development Lonza Personal Care, 70 Tyler Place, South Plainfield, NJ 07080
Introduction
Personal Care is one of the sectors with the highest demand and patent activity for the use of botanicals and extracts for various skin care claims. The growing awareness on green initiatives around reducing biomass waste and continuous improvement in the quality of natural products has also influenced this industry. Fermentation technology is a versatile process that can easily be adapted to an ever-changing market that demands products that are natural, sustainable, produced with minimum waste and are completely traceable in origin. It is the ideal platform technology to promote a controlled environment for growing natural sustainable botanicals. ReGeniStemTM Red Rice (R3) is the product of our expertise in plant sciences and bio-fermentation. Figure 1 details the coalescence of the diverse technologies that comprise the concept of this exciting new product. To create this product we culture meristem cells of Himalayan Red Rice, using the method detailed in Figure 2. These cells are grown in a bioreactor and subjected to an ozone stress which causes them to produce secondary metabolites.
Plant Tissue Culture
Plants are unique compared to animals in that plants are able to regenerate a complete organism from a single cell taken from an older plant. The process of growing such plant cells is referred to as plant tissue culturing and has been known since the early 1900’s. Plant tissue culture is the aseptic culture of cells, tissues, or organs under defined physical and chemical conditions in vitro. Gottlieb Haberlandt first proposed the theoretical idea for plant tissue culture in 19021. He predicted that one could successfully cultivate and regenerate an entire plant from a single or a few vegetative cells. He established the concept of totipotency, where isolated plant cells could be cultivated in nutrient media and then be able to eventually generate a whole plant from one of these individual cells. Research eventually led to the culture of root tips and buds in the 1920s to the 1940s2, 3, 4, 5.
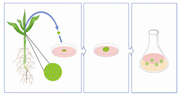
Figure 2. Meristems are taken from the shoot of the plant and transferred to solid culture media for callus growth. Calli of a certain size are then transferred to liquid culture media
One of the first plant tissue cultures was derived from plant tissue containing meristematic cells from Acer pseudoplatanus commonly called Sycamore Maple6. Early studies in the 1930s had shown that cultured root tips were free of viruses7. Morel and Martin confirmed this in 1952 when virus-free Dahlia plants were obtained from infected plants by culturing their shoot tips8. Virus elimination from the meristem is possible because vascular tissues, within which the viruses move, do not extend into the root or shoot apex where the meristem is. The growth of plant cell culturing expanded significantly with the discovery by Murashige and Skoog of a unique nutrient media (now called M&S media) which allowed for the controlled growth of nearly any plant cell in vitro9. The M&S formulation is now the most widely used nutrient medium in plant tissue culture. Currently tissue cultures are being grown with all types of plants, including cereals and grasses, legumes, vegetable crops, oilseeds, temperate and tropical fruits, forest trees, ornamentals, and rare plants.
Secondary Metabolites Induced By Ozone Stress
In their natural environment, plants have to deal with an overabundance of different growing conditions that could either be favorable or detrimental to their growth. Due to their lack of mobility, plants have evolved to produce secondary metabolites in order to overcome these dangers and stressors. Secondary metabolites are such called due to the fact that they do not carry a primary function in the plant such as growth, photosynthesis or reproduction. These secondary metabolites are essential to the well-being of plants but are secondary in terms of their survival. Growing plant cells in a nutrient media does not necessarily make plants produce metabolites similar to those grown in the wild. Often plants grown in nutrient media do not express important secondary plant metabolites because they were not threatened by the atmosphere, adverse soil conditions, or by the presence of pathogens or insects that would damage the plants. However, it was found that certain media components, such as ozone, plant growth regulators, and pathogen polymers such as chitin can cause plants grown in culture to express secondary metabolites10, 11, 12, 13. In particular the use of ozone to cause expression of secondary metabolites and elicit a change in tobacco has been well documented14, 15, 16.
In order for our meristem cultures to produce secondary metabolites, we choose ozone as the stressor, as it is easily introduced into the bioreactor, and ozone was the most effective at causing the culture to produce the most secondary metabolites as compared to other elicitors. It is possible to optimize the amount of ozone applied to the meristem suspension culture in order to achieve specific elicited results.
Product Identification
HPLC (High Performance Liquid Chromatography) analysis was conducted on the meristem conditioned nutrient media exposed to ozone. Although only one peak has been identified at this time as biotin, multiple peaks were observed in the extract, demonstrating that exposure to ozone causes the production of secondary metabolites by the rice meristem culture. These differences are clearly seen in the chromatograms in Figures 3 and 4.
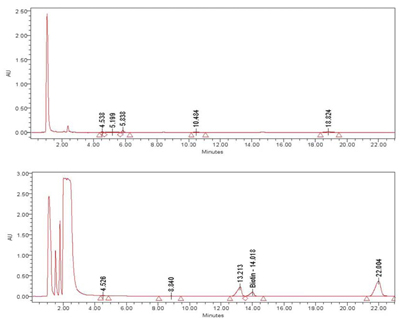
Figure 3. ReGeniStem™ Red Rice without ozone stress; Figure 4. ReGeniStem™ Red Rice with ozone elicitation
Overview of DNA Methylation as a Type of Epigenetic Modification
Epigenetics is the study of heritable changes in gene expression that is caused by a mechanism other than changes to the DNA sequence. Epigenetics’ literal translation is “above the genome”. Thus, any modification that alters gene activity without changing the DNA sequence and that can also be transmitted to daughter cells would be considered an epigenetic modification. Epigenetic changes are passed on to daughter cells and may last for multiple generations even though there is no change to the DNA sequence. Epigenetic factors and changes have been shown to play an important part in cellular differentiation, development, aging and disease17, 18, 19.
Numerous papers have reported on methylation of genes versus the intergenic regions between genes, transcription start sites in the promoter versus genes, and introns versus exons. Further comparisons have looked at differentiated cells versus undifferentiated cells and young differentiated cells versus old differentiated cells. A blanket statement cannot be made that DNA methylation promotes or prevents transcription. The key is where the DNA methylation is occurring. There are many nuances relating to epigenetic modifications and so a detailed examination of the types and patterns of DNA methylation is needed to clarify which epigenetic DNA methylation patterns we are interested in modulating.
DNA CpG Methylation
DNA methylation at both the global and gene specific level has been shown to play an important role in regards to epigenetic memory; changes in DNA methylation have been shown to be age-related20, 21, 22, 23. DNA methylation in regards to epigenetic changes occurs at the cytosine-5 in CpG dinucleotides. The methyl group is attached to a cytosine base followed by a guanine dinucleotide base. The p represents the phosphate that links the C and G together. This helps differentiate CpG from a CG base pair. DNA methylation can also occur at CpA, CpT and CpC sites, although currently the majority of the data linking cytosine methylation and gene transcription is from CpG data due to both biological and technical reasons. There is an estimated 32 million CpG dinucleotides which constitute approximately one percent of the genome24, 25. Other types of epigenetic modifications include changes in chromatin structure through histone acetylation, ubiquitylation, sumolation and methylation26.
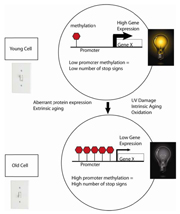
Figure 5. Young cells can be thought of as having their “youth switch” turned on. Young cells have low levels of CpG methylation in the promoter region. As cells age, the promoters become progressively more methylated resulting in diminishment of gene transcription
Methylation of the CpG-rich promoter areas of genes, called CpG islands, has been identified as an essential mechanism in the regulation of gene transcription27, 28. CpG islands have been defined as a region of 200 base pairs with a GC content of >55%, and are in approximately 70% of human promoters24. Besides being associated with promoters, CpG islands are also linked to housekeeping genes. CpG islands are typically unmethylated in normal cells, thus allowing the transcriptional machinery access to the DNA. In certain diseases and with aging, CpG islands become aberrantly methylated, deregulating the usual transcriptional activity of the gene. The methylation occurs symmetrically on the cytosine residues on both strands of the CpG dinucleotides. When CpG islands in a promoter are highly methylated, this results in transcriptional repression of the adjacent gene. Expressed genes generally appear to have low methylation in their promoter region and high methylation in their gene body29. Young cells can be thought of as having their “youth switch” turned on. At the cellular level, this translates to having a low level of CpG promoter methylation, allowing high gene expression. As a cell ages, the “youth switch” is turned off. The promoter accumulates higher levels of methylation, decreasing gene transcription (Figure 5). DNA methylation is presumed to change the chromatin density and accessibility of the DNA to cellular machinery, thus influencing the transcriptional potential of the underlying DNA sequence.
Global and Gene Promoter DNA Methylation
DNA methylation is established early in an organism during embryogenesis; DNA hypermethylation is the prime mechanism of X-chromosomal inactivation30. Differentiation is associated with a more restricted pattern of gene expression31. As the cells transition from an undifferentiated state to a differentiated one, CpG methylation patterns help to set up the pattern of gene expression. In an undifferentiated state, the cell hasn’t committed to being a certain type of cell and so most of its genes have the potential to be expressed. As differentiation occurs, gene expression patterns are determined with certain genes being expressed and other genes being turned off. DNA methylation helps to control this pattern of gene expression (Figure 6).
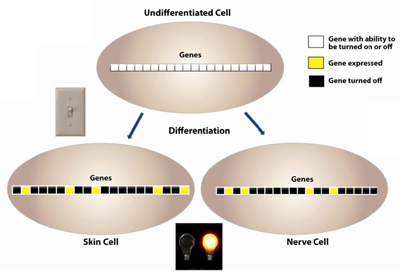
Figure 6. As the cells transition from an undifferentiated state to a differentiated one, CpG methylation patterns help to set up the pattern of gene expression. In an undifferentiated state, the cell hasn’t committed to being a certain type of cell and so most of its genes have the potential to be expressed. As differentiation occurs, gene expression patterns are determined with certain genes being expressed and other genes being turned off. DNA methylation helps to control this pattern of gene expression
Epigenetic Changes and Aging
Aging is influenced by a multitude of factors including oxidative damage to DNA, accumulation of DNA mutations, telomere shortening, senescence, apoptosis, mitochondrial mutations, accumulation of intracellular junk and extracellular junk and extracellular cross linking. Deteriorations of the epigenetic marks lead to impaired cellular and molecular functioning. Hypermethylation is associated with heterochromatin and transcriptional silencing. Perturbances in DNA methylation patterns can be associated with malfunctions of cancer cells’ gene expression, especially down-regulation of genes with tumor suppressor functions through hypermethylation of their promoter regions32.
CpG islands gain methylation with age while CpG sites not in CpG islands lose methylation with age. The CpG islands of many promoters that are usually unmethylated become methylated33, 34. Hence in younger cells, the CpGs located in islands tend to be unmethylated. For humans, promoters tend to gain methylation with age. This trend can be seen when viewing the variation in methylation in regards to aging and environmental exposure to toxins such as exposure to smoking, arsenic or asbestos35, 36, 37, 38, 39.
DNA Methylation Summary and Experiment Introduction
Epigenetics can be thought of as a “youth switch”. The switch is turned on in young cells and turned off as cells age. DNA methylation is a component of this youth switch and DNA methylation could be considered as a stop sign in gene promoters. Young healthy cells are thought to maintain their CpG islands in a low methylated state to allow transcription to occur. Less methylation is fewer stop signs to prevent the gene from being expressed. In a young cell, there is a low amount of promoter DNA methylation, leading to high gene expression and thus high protein production. As aging occurs and DNA methylation drift increases the amount of DNA promoter methylation, the gene is expressed less causing lower protein production. Our hypothesis was that undifferentiated plant cells may help reverse promoter methylation. As mentioned above, undifferentiated cells have different CpG methylation patterns compared to differentiated cells, and their promoter DNA methylation levels are lower. There may be various plant transcription factors, cellular components and homologous proteins and peptides that may be able to be applied to aged skin and help the skin cells to rejuvenate by decreasing promoter DNA methylation (Figure 7). By profiling the CpG methylation differences in young and aged skin tissues, characterization of the role of aging related to methylation variation can be observed. The following studies were done to show the effect of ReGeniStemTM Red Rice on aged cells focusing on these important sites of methylation. The changes in DNA methylation patterns will manifest as changes in critical protein expression, making aged cells behave more like young cells.
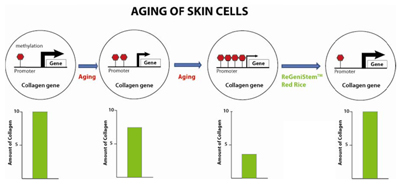
Figure 7
Global Decrease of CpG Methylation in Gene Promoter Regions
ReGeniStemTM Red Rice was evaluated using the DNA CpG methylation chip. The DNA CpG Methylation Array is a new DNA microarray based platform for studying methylated DNA that is specifically focused on CpG sites and identifies approximately 250,000 CpG sites in the human genome. It is a highly specific tool that allows an accurate measurement of methylated sites for better understanding of aging, disease and the identification of epigenetic markers (Agilent Technologies).
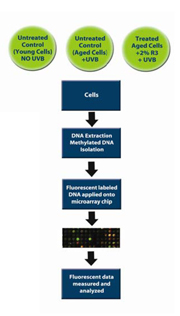
Figure 8. Schematic diagram of the in vitro test methodology used to analyze ReGeniStem™ Red Rice’s effect on DNA methylation reduction
The evaluation of global CpG methylated sites involved a two-step process wherein three sets of human dermal fibroblast cultures were intrinsically (cell culture passage) and extrinsically (UVB light exposure) aged. These cells were divided into three groups and then treated as follows:
A. Control (Young Cells): untreated cells, no UVB
B. Control (Old Cells): untreated cells, extrinsically and intrinsically aged
C. Treated Cells: treated with 2% ReGeniStemTM Red Rice, extrinsically and intrinsically aged
At the end of the treatment period, the genomic DNA was extracted from each group of fibroblasts and placed into separate solutions that underwent a number of steps that eventually lead to the fluorescent labeling of the methylated DNA groups. The labeled DNA samples were then placed in the DNA methylation microarray and analyzed (Figure 8). The resulting data consisted of three sets of approximately 250,000 CpG sites each. Since we were interested in the change of methylation at the promoter level, all CpG sites within all promoters were analyzed (Graph 1). As expected, there is an increase of global methylation at the promoter level when comparing young cells to old cells, both without ReGeniStem Red Rice. The application of 2% ReGeniStem Red Rice to old cells had the ability to modulate the epigenetic environment and decrease the global level of promoter methylation, perhaps helping the old cells to function and perform as a younger, healthier cell.
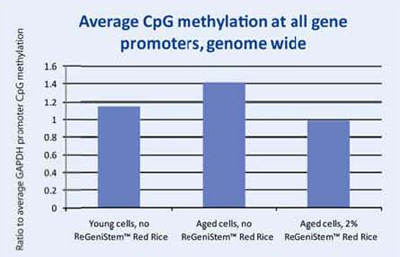
Graph 1. Average Global CpG methylation at the promoter region with ReGeniStem™ Red Rice treatment
CpG Methylation at Collagen 1A1 (COL1A1) Gene
When a specific gene, COL1A1, was examined for its promoter methylation levels, the resulting data showed that treatment with 2% ReGeniStemTM Red Rice also reduces CpG methylation at the COL1A1 promoter region on treated aged cells (Graph 2). As with the global promoter methylation data trend, there is an increase in CpG promoter methylation when comparing young and old cells untreated with ReGeniStem Red Rice. The decrease in promoter methylation of the Col1A1 gene from application of ReGeniStemTM Red Rice should help to increase Col1A1 gene expression and hence protein production.
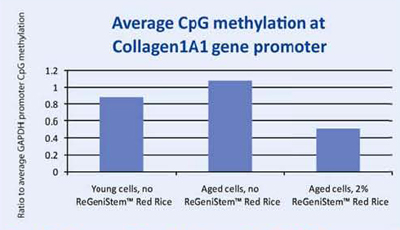
Graph 2. Average CpG methylation at the COL1A1 gene promoter
In Vitro Pro-collagen 1 Synthesis
To correlate DNA demethylation to Collagen 1A synthesis, a protein assay was conducted on human fibroblasts treated with and without 2% ReGeniStemTM Red Rice. The results shown in Graph 3 confirm that treatment with 2% ReGeniStemTM Red Rice increases collagen synthesis in cells when compared to old untreated cells.
Dermatopontin Expression in Keratinocytes
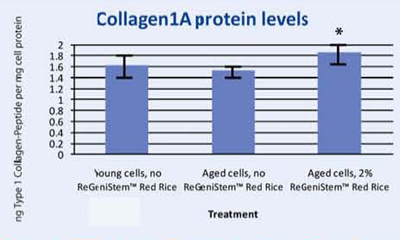
Graph 3. Protein assay showing the effect of ReGeniStem™ Red Rice on collagen synthesis
Dermatopontin is a protein found in the extracellular matrix of the dermis. It has been known to have multiple and diverse functions in the cells; ranging from accelerating collagen fibrillogenesis to fibroblast and neurogenic cell adhesion40. Recent studies have shown that dermatopontin (DP) may play a role in controlling the organization of collagen fibrils in the dermis. Another study suggests that dermatopontin is responsible for epidermal adhesion of keratinocytes to the epidermis41. Our microarray assay uncovered that this gene was upregulated in keratinocytes treated with ReGeniStemTM Red Rice. We then assayed dermatopontin protein expression in keratinocytes that were either untreated or treated with 1% or 2% ReGeniStemTM Red Rice. Dermatopontin was upregulated in the 2% treated tissue. This suggests that ReGeniStemTM Red Rice may be able to firm the skin and influence the skin barrier function especially in the dermal and epidermal layers where this protein is found. This finding that dermatopontin is expressed in keratinocytes and can be upregulated is also in itself a novel finding since no one in the scientific literature has previously published that dermatopontin is expressed in keratinocytes.
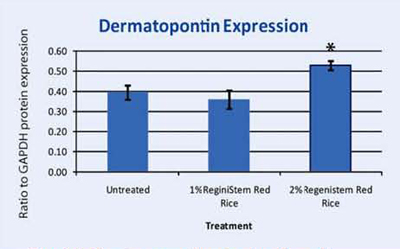
Graph 4. Protein assay showing the effect of ReGeniStem™ Red Rice on dermatopontin synthesis
Conclusion
Growing ReGeniStem™ Red Rice in bioreactors is a sustainable process that reduces residual biomass waste, but the benefits of this technology are not limited to environmental impact. Through elicitation of our red rice meristematic cell cultures, we were able to produce appreciable quantities of secondary metabolites in the culture. When the culture was tested on fibroblasts in vitro, we show a reverse in the age related increase of methylation in the promoter region of genes. This rejuvenation at the epigenetic level may have multiple benefits to the cells and to the skin. Some benefits include anti-aging, the renewal of gene and protein expression levels to that found in younger cells, such as was shown for collagen and the reversal of age related decreases in protein production. In essence, the skin, through an application of ReGeniStemTM Red Rice, is pushed to a younger, healthier state. An increased expression of dermatopontin in keratinocytes, besides being a novel finding, implies that ReGeniStemTM Red Rice may be able to firm the skin and improve the skin barrier function.References
1. Haberlandt G (1902) Kulturversuche mit isolierten Pflanzenzellen. Sitzungsber. Akad. Wiss. Wien. Math.-Naturwiss. Kl., Abt. J. 111, 69–92
2. Kotte W (1922) Wurzelmeristem in Gewebekultur. Bmichte der deutsche botanische Gesellrchaj 40: 269-272
3. Robbins WJ (1922) Cultivation of excised root tips and shoot tips under sterile conditions. Botanical Gaze& 73: 376–390
4. Loo SW (1945) Cultivation of excised stem tips of Asparagus in vitro. American Journal Botany 32: 13-17
5. Ball JX (1946) Developments in sterile culture of stem tips and subjacent regions of Tropaeolum majus L. and of Lupinus albus L. American Journal of Botany 33: 301–318
6. Gautheret RJ (1935) Rechmchs mr la culture des tissus udgdtaux. Essais de culture de qulques hsus mhisthatiques. Paris: Le Francois
7. White PR (1934) Multiplication of viruses of tobacco and Aucuba mosaics in growing excised tomato root tips. Phytopathology 24: 1003-1011
8. Morel G, Champagnat M (1969) Diffkrent modes d'kvolution de I'apex du cours de la formation de protocormes. Proceedings of the 2nd European Orchid Congress, Paris, 20-26
9. Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15: 473-497
10. Zenk, M H (1978) The impact of plant cell culture on industry. In T. A. Thorpe (Ed.), Frontiers of plant tissue culture (pp.1–13). Intl. Assoc. Plant Tissue Culture, Univ. of Calgary Printing Services.
11. Wink M (1987) Physiology of the accumulation of secondary metabolites with special reference to alkaloids. In F. Constabel & I. K. Vasil (Eds.), Cell culture and somatic cell genetics of plants (Vol. 4, pp. 17–42). New York: Academic Press.
12. Kurz WGW (1988) Semicontinuous metabolite production through repeated elicitation of plant cell cultures: A novel process. In T. J. Mabry (Ed.), Plant biotechnology (pp. 93–103). Austin: IC2 Institute.
13. Alonso R, Elvira S, Castillo F J, Gimeno B S (2001) Interactive effects of ozone and drought stress on pigments and activities of antioxidative enzymes in Pinus halepensis. Plant Cell Envirn 24(9): 905-916
14. Keinanen M, Oldham N, Baldwin I T (2001) Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuate. J Agric Food Chem. 49 (8): 3553-3558
15. Janzik I, Preiskowski S, Kneifel H (2005) Ozone has dramatic effects on the regulation of the prechorismate pathway in tobacco (Nicotiana tabacum L. cv. Bel W3). Planta 223: 20-27
16. Lee T T (1968) Effect of ozone on swelling of tobacco mitochondria. Plant Phys 43(2) 133-139
17. Cropley JE, Suter CM, Beckman KB, Martin DI (2006) Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc Natl Acad Sci U S A 103: 17308–17312
18. Weaver IC, Meaney MJ, Szyf M (2006) Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A 103: 3480–3485
19. Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, et al. (2003) Transgenerational inheritance of epigenetic states at the murine Axin (Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A 100: 2538–2543
20. Gopisetty G, Ramachandran K, Singal R (2006) DNA methylation and apoptosis. Mol Immunol 43: 1729–1740
21. Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, et al. (1994) Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res 54: 2552–2555
22. Feng YQ, Desprat R, Fu H, Olivier E, Lin CM, et al. (2006) DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet 2: e65
23. Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA (2009) The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 4(8):e6767
24. Saxonov S, Berg P, Brutlag DL (2006) A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA 103: 1412–1417
25. Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, et al. (1982) Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 10: 2709–2721
26. Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128: 635–638
27. Metivier R, Gallais R, Tiffoche C, Le PC, Jurkowska RZ, et al. (2008) Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50
28. Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21
29. Ball MP, Li JB, Ago Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM (2009) Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27(4): 361-368
30. Razin A, Shemer R (1995) DNA methylation in early development. Hum Mol Genet 4 Spec No: 1751–1755
31. Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Sung KWK, Rigoutsos I, Loring J, Wei CL (2010) Dynamic changes in the human methylome during differentiation. Genome Res 20: 320-331
32. Ting AH, McGarvey KM, Baylin SB. (2006) The cancer epigenome components and functional correlates. Genes & Dev 20: 3215–3231
33. So K, Tamura G, Honda T, Homma N, Wai T, Togawa N, Nishizuka S, Motoyama T (2006). Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer Sci 97: 1155-1158
34. Waki T, Tamura G, Sato M, Motoyama T (2003) Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene 22: 4128-4133
35. Christensen BC, Godleski JJ, Marsit CJ, Houseman EA, Lopez-Fagundo CY, et al. (2008) Asbestos exposure predicts cell cycle control gene promoter methylation in pleural mesothelioma. Carcinogenesis 29: 1555–1559
36. Lyon CM, Klinge DM, Liechty KC, Gentry FD, March TH, et al. (2007) Radiation-induced lung denocarcinoma is associated with increased frequency of genes inactivated by promoter hypermethylation. Radiat Res 168: 409–414
37. Marsit CJ, Houseman EA, Schned AR, Karagas MR, Kelsey KT (2007) Promoter hypermethylation is associated with current smoking, age, gender and survival in bladder cancer. Carcinogenesis 28: 1745–1751
38. Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, et al. (2003) Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer 103: 153–160
39. Marsit CJ, McClean MD, Furniss CS, Kelsey KT (2006) Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer 119: 1761–1766
40. Forbes EG, Cronshaw AD, MacBeath JR, Hulmes DJ (1994) Tyrosine-rich acidic matrix protein (TRAMP) is a tyrosine-sulphated and widely distributed protein of the extracellular matrix. FEBS Lett 351(3): 433-436
41. Okamoto O, Hozumi K, Katagiri F, Takahashi N, Sumiyoshi H, Matsuo N, Yoshioka H, Nomizu M, Fujiwara S (2010) Dermatopontin promotes epidermal keratinocyte adhesion via α3β1 integrin and a proteoglycan receptor. Biochemistry 49: 147-15
